Group leader
José-Luis Vazquez-Ibar
JoseLuis.VAZQUEZ-IBAR@cea.fr
Amino acid transporters
Amino acid transporters are
essentials players in human physiology as they participate on protein
synthesis, energy metabolism, gene expression, redox balance or signal
transduction pathways. One of the largest families of amino acid transporters
is the L-amino acid transporter (LAT) family, where congenital mutatios of two
members of this family cause two pathological disorders: cystinuria and
lysinuric protein intolerance. In collaboration with Prof Manuel Palacín (IRB,
Barcelona, Spain) we solved the X-ray structure of a LAT prokaryotic homolog:
Adic (Kowalczyk, L
et al, 2011, Figure 1), revealing for the first time an induced-fit
mechanism during the first step of substrate-protein interaction. While two
structures of mammalian LATs have recently appeared, prokaryotic LATs are still
very valuable tools to continue deciphering the transport mechanism and
regulation of their mammalian homologs. Using a directed evolution strategy
(Rodriguez-Banqueri et al 2016), we have generated engineered versions of SteT
(a prokaryotic paradigm of LATs) trapped in different stages along the
catalytic cycle. Structure-function analyses of these mutants are expected to
improve our understanding of the transport mechanism and lipid regulation of
LATs.
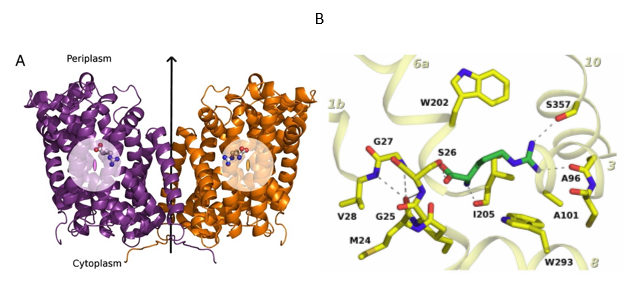
Figure 1: Structure
of a mutated version of Adic bound to the substrate, Arg+. (A)
The transporter forms a homodimer (in purple, protomer 1 and in orange,
protomer 2). The bound substrate is depicted with a ball-and-stick model. (B) Detail of the binding site. The
structure shows the delocalization of the substrate in the binding site as a
consequence of removing one hydrogen bond interaction between the substrate and
the transporter, providing structural evidences of a substrate’s induce fit
mechanism of these transporters.
In collaboration with Dr Bruno Gasnier (Paris-Descartes) and Dr Liang Feng (Stanford University, USA), we are also studying the lysosomal cationic amino acid transporter PQLC2. PQLC2 activity is likely to be linked with mTORC1 signaling by tightly controlling intralysosomal arginine levels. In addition, PQLC2 mediates the recruitment of the C9orf72 complex to the lysosomal surface. Mutations in the gene encoding C9orf72 causes amyotrophic lateral sclerosis and frontotemporal dementia. Our aims include unraveling the transport mechanism and regulation of PQLC2 as well as the study of the structural and functional consequences of PQLC2 interaction with the C9orf72 complex.