Neuroimaging-genetic cohorts in general population bring together two
types of data: brain imaging and genetic data. These are unique and valuable
tools for discovering associations between genetic variants and characteristics
derived from brain imaging (concept of endophenotypes*). They are also
invaluable resources for studying the relative influence of genetics and
environment on the variance of brain characteristics observed in normal and
pathological populations. Analysis of such cohorts allows the search for
genetic associations with earlier or more reliable characteristics than the
classic binary phenotypes, such as diagnostics observable in an individual.
The UK Biobank general population cohort, which contained 16,304 subjects at
the time of the study, is currently the best example in the world of the use
that can be made of this type of tool. This cohort allowed the investigators to
conduct a genome-wide haplotype analysis (sequences of multiple contiguous
variants inherited from the same parent) for the opening value of 123 brain
sulci (sulci are the elongated depressions observed on the surface of the cortex).
A first originality of the work is that association studies are usually
conducted for each individual genetic variant and, when haplotypic studies are
conducted, they are performed on very limited portions of the genome, for
reasons of computational burden. The second originality is the endophenotype
analyzed, i.e. the opening of the sulci distributed throughout the cortex,
which is an indicator of brain aging.
Using genetic maps, the researchers defined 119,548 low-recombination blocks
distributed along the 22 autosomal chromosomes and analyzed 1,051,316
haplotypes. They thus confirmed the genetic particularity of the regulatory
region of the KCNK2 gene located on chromosome 1 and its association with the
opening of the sulci of the cingulate cortex. They also identified three
additional genomic regions of interest (on chromosomes 9, 12, and 16), whose
found association is independently replicated in approximately 5000 additional
subjects.
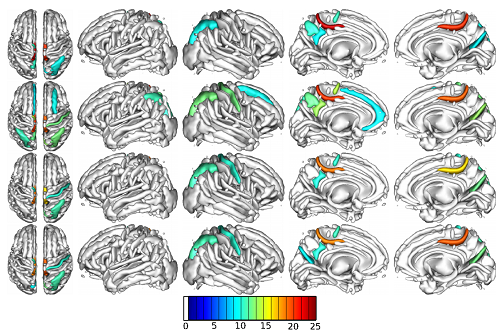
Interior volumes of cortical sulci whose opening was studied. Colored sulci are those found associated with a haplotypic region of chromosome 1. Color code: level of significance of association.
© S Karkar, E Le Floch, V Frouin /CEA
This work has made it possible to map sulci that show variability in opening that can be partially explained by regions of the genome. They also allowed to compare the relative interests of classical association methods with the haplotypic approach chosen here, showing in particular that genome-wide haplotype analyses are more sensitive than SNP** (Single Nucleotide Polymorphism) approaches and represent an interesting and statistically robust alternative.
Contact : Vincent Frouin
* The concept of endophenotype was
created to categorize complex, primarily mental, disorders into simpler,
stable, intermediate manifestations accurately measured by neuroimaging, that
have a proven genetic link to disease.
** SNPs (Single Nucleotide Polymorphism)
are the most abundant form of genetic variation in the human genome. They
account for more than 90% of all differences between individuals. It is a type
of DNA polymorphism in which two chromosomes differ on a given segment by a
single base pair.