Skills and tools
The LSOD includes researchers characterizing the molecular mechanisms of stress-related proteins (hemoproteins, flavoproteins and metalloproteins) or detoxification (cytochromes P450, glutathione S-transferase, P-glycoprotein), using biochemical, enzymological, biophysical and bioinformatics techniques.
- Molecular biology for the production of native and mutant proteins (heterologous expression systems: Escherichia coli, Lactococcus lactis).
- Biochemical methods of production, identification and characterization of proteins and / or protein domains, substrates and metabolites.
- Contribution from chemistry: synthetic and biomimetic models (heme and non heme, copper and manganese complexes).
- Raman, resonance Raman and FTIR techniques for characterizing the structure and environment of the chromophores (heme, flavins, ...) studied.
- Electron Paramagnetic Resonance spectroscopies (EPR, ENDOR, ESEEM, HYSCORE) that allow the structural characterization of paramagnetic species (radicals, CuII, FeIII, ...).
- Rapid kinetics (stopped-flow, freeze-quench) and catalytic production (fluorimetry, HPLC ...) methods.
- Molecular modelling methods.
Detoxification and oxidative stress
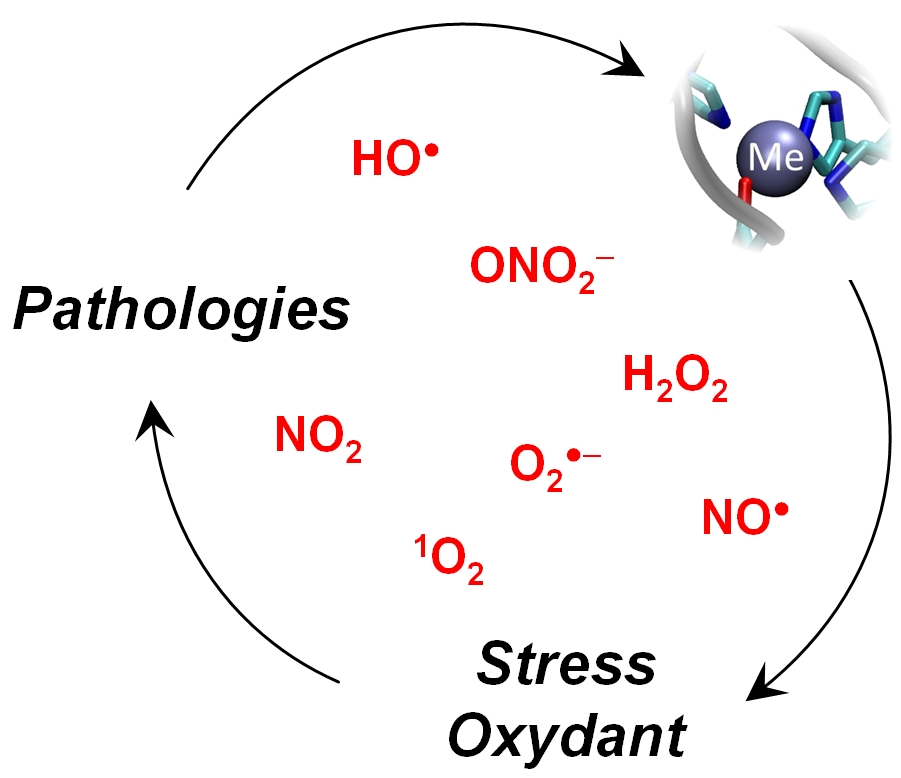 Oxidative stress is an essential phenomenon to the initiation and development of many diseases including cancer, inflammatory diseases, cardiovascular and neurodegenerative diseases. However, the mechanisms of detoxification and production of reactive oxygen and nitrogen species (RNOS see Figure) and their biological reactivity are not yet characterized in a clear and complete fashion. This is illustrated in the case of neurodegenerative diseases for which the presence and activity of detoxification enzymes have been described only recently and partially. RNOS seem to play different if not opposite roles, such as signalling, neuroprotection or cytotoxicity. Our team strives to decipher the catalytic processes of detoxification or production of RNOS, with special attention to several systems strongly implicated in these diseases :
Oxidative stress is an essential phenomenon to the initiation and development of many diseases including cancer, inflammatory diseases, cardiovascular and neurodegenerative diseases. However, the mechanisms of detoxification and production of reactive oxygen and nitrogen species (RNOS see Figure) and their biological reactivity are not yet characterized in a clear and complete fashion. This is illustrated in the case of neurodegenerative diseases for which the presence and activity of detoxification enzymes have been described only recently and partially. RNOS seem to play different if not opposite roles, such as signalling, neuroprotection or cytotoxicity. Our team strives to decipher the catalytic processes of detoxification or production of RNOS, with special attention to several systems strongly implicated in these diseases :
- Metallo-peptides such as Cu-amyloid b involved in Alzheimer's disease. A key aspect of their toxicity is linked to the production of ROS
- NO synthases (NOS) are now regarded as playing a key role in several neurodegenerative and cardiovascular diseases through the production of RNOS
- Development of new agents active against cell lines representative of medulloblastoma and their mechanisms of action towards cytochromes P450 and production of RNOS
Xenobiotic metabolism
To fight against aggressions by xenobiotics, the human body has developed three specialized and complementary enzyme systems for the elimination of foreign compounds and which are involved in cellular detoxification (see Figure). These enzymes are cytochromes P450 (CYPs) in the phase functionalization (Phase I metabolism of xenobiotics), conjugation enzymes so-called transferases (phase II) such as the glucuro- or glutathione-S-transferases (including membrane mGST1), and transport proteins such as efflux and P-glycoprotein (P-gp) (Phase III). They are localized in different membranes (endoplasmic reticulum or plasma membrane) and can act synergistically to produce efficient detoxifications of the liver, the principal organ exposed to xenobiotics by the oral route. CYP3A4 and P-gp are characterized by a multispecific molecular recognition towards a great variety of chemical structures and share many common substrates. Because of their hydrophobicity, these common substrates are recognized by these enzymes within the membrane.
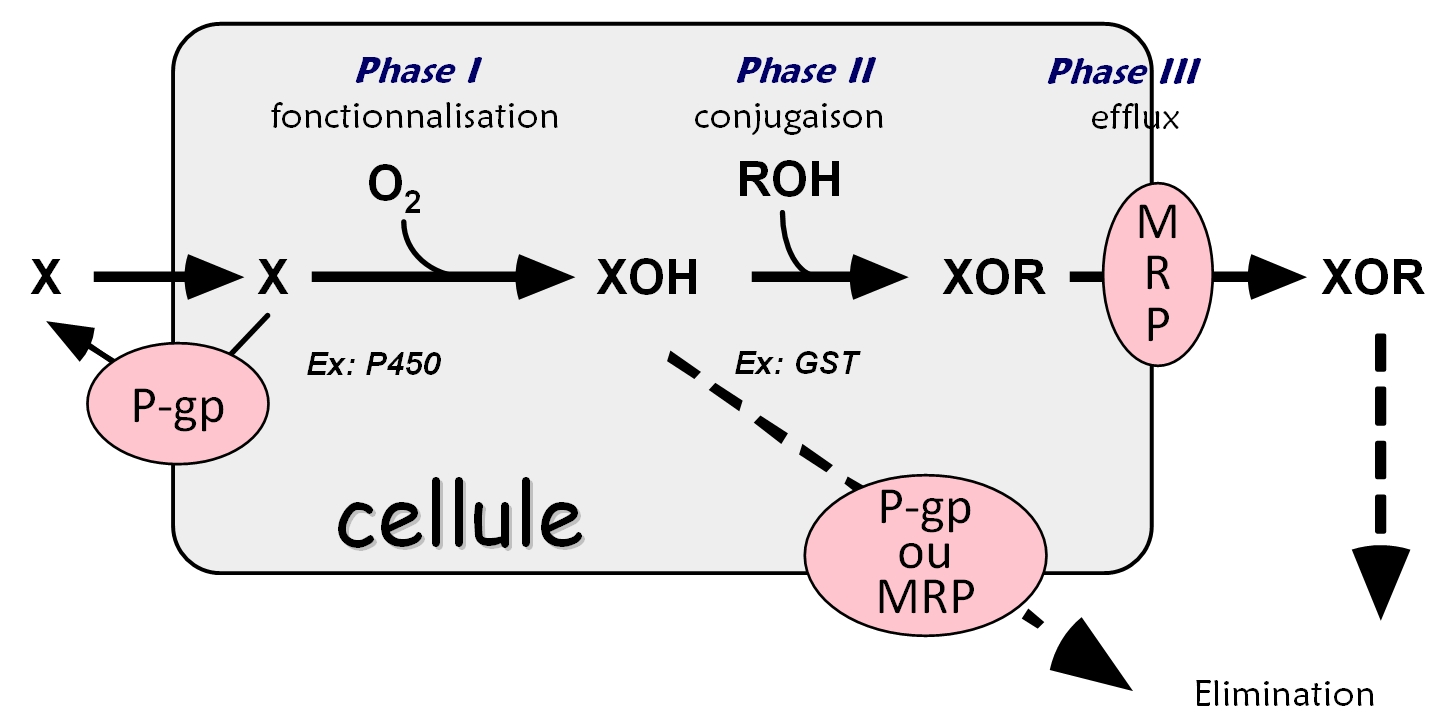
We are interested in understanding the functional synergy of the undertaking by mGST1 or P-gp of metabolites produced by P450s, particularly CYP3A4, and in determining the molecular mechanisms of multispecies recognition of these three membrane proteins. These enzymes are thus able to operate "in parallel" to protect the cell against hydrophobic toxic species but also "in series", that is, a product of the enzymatic activity of CYP3A4 could be transported out of the cell by P-gp or via mGST1.
The investigated molecules are in particular drugs in development and compounds found in our environment and that can lead to health problems.
Structure-function studies of oxidative stress enzymes.
At a molecular level, we study the structure-function relationships of a number of proteins that play an important role in oxidative stress and cellular detoxification. To better understand the catalytic mechanisms and the intramolecular electron (and possibly proton) transfer at the origin of the functional properties of these proteins, we aim in particular at determining the structure and environment heme of active sites (NOS, NOS-like proteins, cytochromes P450, oxygen carriers, amyloid β-heme complex), non-heme centers (metal-peptide complexes, superoxide reductase) and flavin (flavoproteins belonging to the pyridine nucleotide-disulfide oxidoreductases family , NOS reductase domain).
A wide range of methodologies is implemented. It includes enzymology, molecular biology, rapid kinetics measurements, biomimetic chemistry, vibrational spectroscopies (IR and Raman), electronic absorption and electron spin resonance (conventional and pulsed EPR, ENDOR), and bioinformatics (molecular dynamics, docking).