Three main techniques are involved in the analysis facilities of SIMOPRO: Amino Acids Analysis, Edman Sequencing and Mass Spectrometry. They are used for both the quality control of all peptides and proteins production and for technically supporting SIMOPRO’s research projects.
Team Leader
Robert THAI
01 69 08 99 61
robert.thai@cea.fr
Routine analyses
Routine analyses for quality control and molecular characterization of all molecules produced and studied in SIMOPRO are performed on:
- Aminoacids Analyzer 'AminoTac' from JEOL for routine quantification of peptide/protein samples ;
- Edman sequencer 'Procise 492HT' from Applied Biosystems for identifying the N-terminal sequence of peptide/protein and particularly for
de novo sequencing of new compounds;
- 2 Mass Spectrometers ESI-MS ion-Trap 'Esquire HCT' from Bruker-Daltonics and 'MALDI-TOF/TOF 4800' from AB SCIEX for validating molecules integrity and their structural characterization.
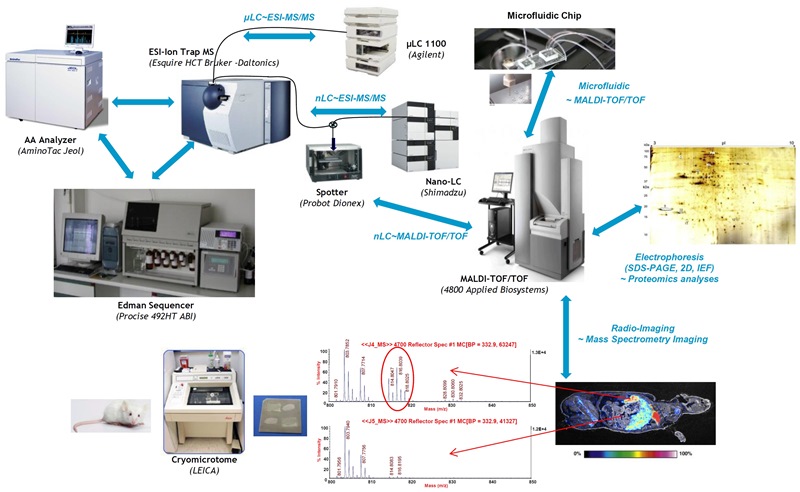
Mass Spectrometry
The last technique, Mass Spectrometry, has been rapidly growing for the last few years and becomes inescapable to technically support all SIMOPRO's research projects:
- by peptide mapping methodology to identify and localize chemical modifications in engineered proteins, including the isolation of a reaction intermediate in an enzymatic pathway (MALDI-TOF/TOF);
- by identifying potential high value-added natural peptide molecules for new biosynthesis pathways (LC~ESI-MS/MS);
- by sustaining chemical projects based on selective capture of proteins biomarkers in complex proteomes by chemical probes (2D Electropheris, IEF, nLC-ESI-MS/MS et nLC-MALDI-TOF/TOF) including the conception of an innovative coupling of microfluidic technology to MALDI-TOF/TOF;
- by identifying proteins specifically associated with Carbon Nanotubes in biological extracts via preliminary 2D-gels (MALDI-TOF/TOF);
- by identifying naturally processed immunogenic protein sequences (T-epitopes) directly from MHC complexes isolated from APCs (nLC-MALDI-TOF/TOF);
- by screening designed selective inhibitors of metalloproteases including MMPs, Botulinum toxin or Yersinia pestis MCP (LC-ESI-MS/MS) ;
- by screening new specific tracers (Tc) for molecular imaging and development of new proteic ligands used as specific probes in molecular imaging studies (LC~ESI-MS/MS) ;
- by confirming the structure of radiolabelled compounds detected in tissues sections by b–Imager in Imaging Mass Spectrometry methodology.