Recombining for better mixing
During meiosis, recombination of genetic material between homologous chromosomes occurs, which contributes to genetic mixing. Recombination can only take place through fine mechanisms of breakage and subsequent repair of DNA molecules. These mechanisms are highly conserved in eukaryotes, from yeast to humans. In particular, cross-shaped structures (Holliday junctions) are formed between homologous chromosomes to make crossovers during recombination. These recombination mechanisms are essential to ensure correct segregation of chromosomes in gametes. Failure of these steps results in the generation of gametes with an abnormal number of chromosomes (trisomy, Turner syndrome, infertility).
Cut, repair... with what specificity?
The Mlh1-Mlh3 complex is a DNA mismatch repair factor that are generated as a result of errors in replicative polymerases. Mlh1-Mlh3 has endonuclease activity, i.e. it can cut a DNA molecule between two successive nucleotides and not at its ends. Unlike other similar mismatch repair factors, Mlh1-Mlh3 also exerts its endonuclease activity during meiosis, at the Holliday junctions; it is essential for the exchange of chromosome portions. The Mlh1-Pms1 complex, which is the main factor for repairing DNA mismatches, is not involved in meiosis. However, the two complexes have many structural similarities. For example, they are formed by the interaction between the C-terminal domains of Mlh1 and of its partner Mlh3 or Pms1. What are the structural bases for the differences in specificity between Mlh1-Mlh3 and Mlh1-Pms1? Researchers from CEA-Joliot (I2BC department, Structural Biology Laboratory) and the Institut Curie (Team Chromosome dynamics and recombination), with the help of CEA-Jacob (IRCM department) and a Swiss team, have solved the three-dimensional structure of a complex formed by the interaction domains of Mlh1 and Mlh3 (from purified recombinant proteins) of the yeast S. cerevisiae by radiocrystallography, and have functionally characterized it. They compared it with the already known equivalent complex formed between Mlh1 and Pms1.
Their study, published in PNAS, reveals differences between the two complexes, particularly with regard to the size of the heterodimerisation interface. The regulatory domains of Pms1 and Mlh3 are oriented differently in the complex with Mlh1. In addition, the shape of the cavity surrounding the endonuclease site varies, which could lead to differences in specificity towards their DNA substrates. The last ten residues of Mlh1 are known to be essential for mismatch repair but not for interaction with Pms1 or Mlh3. Experiments with mutant S. cerevisiae strains - into which deletions of the last residues of Mlh1 have been introduced - indicate that only the last 3 residues are essential for the meiotic activity of Mlh1. Other (DNA binding) experiments allow the authors to conclude that Mlh1-Mlh3, but not Mlh1-Pms1, binds preferentially to Holliday junctions. Finally, in the crystal, the Mlh1-Mlh3 dimers associate with each other to form a filamentary structure. This conformation supports the hypothesis proposed in previous studies that Mlh1-Mlh3 is oligomerised along the DNA. Mutations in the corresponding interaction surfaces strongly decrease the formation of chromosome crossovers. The authors of the study propose that Mlh1-Mlh3 oligomerization starts at a Holliday junction before extending away along the DNA.
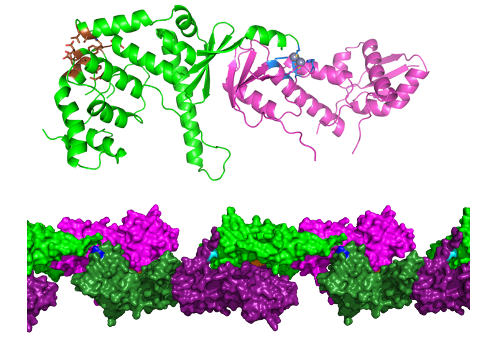
Crystallographic structure of the C-terminal region Mlh1-Mlh3 with its endonuclease site (blue) and its interaction site with meiotic recombination or DNA repair factors (brown). Mlh1-Mlh3 forms oligomers, a possible arrangement of which is observed in the crystal. © JB. Charbonnier/CEA
This first comparison at the structural level of the Mlh1-Mlh3 and Mlh1-Pms1 complexes strongly suggests an evolutionary specialization of each. To go further, it will be necessary to obtain cryo-electron microscopy structures of the two whole proteins, in interaction with their DNA substrate.
Contact @Joliot :
Jean-Baptiste Charbonnier (jb.charbonnier@cea.fr)